We’ve finally reached the end of the road. This is the fifth and last post in a series building up to a Bayesian proportional hazards model for analyzing a stepped-wedge cluster-randomized trial. If you are just joining in, you may want to start at the beginning.
The model presented here integrates non-linear time trends and cluster-specific random effects—elements we’ve previously explored in isolation. There’s nothing fundamentally new in this post; it brings everything together. Given that the groundwork has already been laid, I’ll keep the commentary brief and focus on providing the code.
Simulating data from a stepped-wedge CRT
I’ll generate a single data set for 25 sites, each site enrolling study participants over a 30-month period. Sites will transition from control to intervention sequentially, with one new site starting each month. Each site will enroll 25 patients each month.
The outcome () is the number of days to an event. The treatment () reduces the time to event. Survival times also depend on the enrollment month—an effect I’ve exaggerated for illustration. Additionally, each site has a site-specific effect , which influences the time to event among its participants.
Here are the libraries needed for the code shown here:
library(simstudy)
library(data.table)
library(splines)
library(splines2)
library(survival)
library(survminer)
library(coxme)
library(cmdstanr)Definitions
def <- defData(varname = "b", formula = 0, variance = 0.5^2)
defS <-
defSurv(
varname = "eventTime",
formula =
"..int + ..delta_f * A + ..beta_1 * k + ..beta_2 * k^2 + ..beta_3 * k^3 + b",
shape = 0.30) |>
defSurv(varname = "censorTime", formula = -11.3, shape = 0.36)Parameters
int <- -11.6
delta_f <- 0.80
beta_1 <- 0.05
beta_2 <- -0.025
beta_3 <- 0.001Data generation
set.seed(28271)
### Site level data
ds <- genData(25, def, id = "site")
ds <- addPeriods(ds, 30, "site", perName = "k")
# Each site has a unique starting point, site 1 starts period 3, site 2 period 4, etc.
ds <- trtStepWedge(ds, "site", nWaves = 25,
lenWaves = 1, startPer = 3,
grpName = "A", perName = "k")
### Individual level data
dd <- genCluster(ds, "timeID", numIndsVar = 25, level1ID = "id")
dd <- genSurv(dd, defS, timeName = "Y", censorName = "censorTime", digits = 0,
eventName = "event", typeName = "eventType")
### Final observed data set
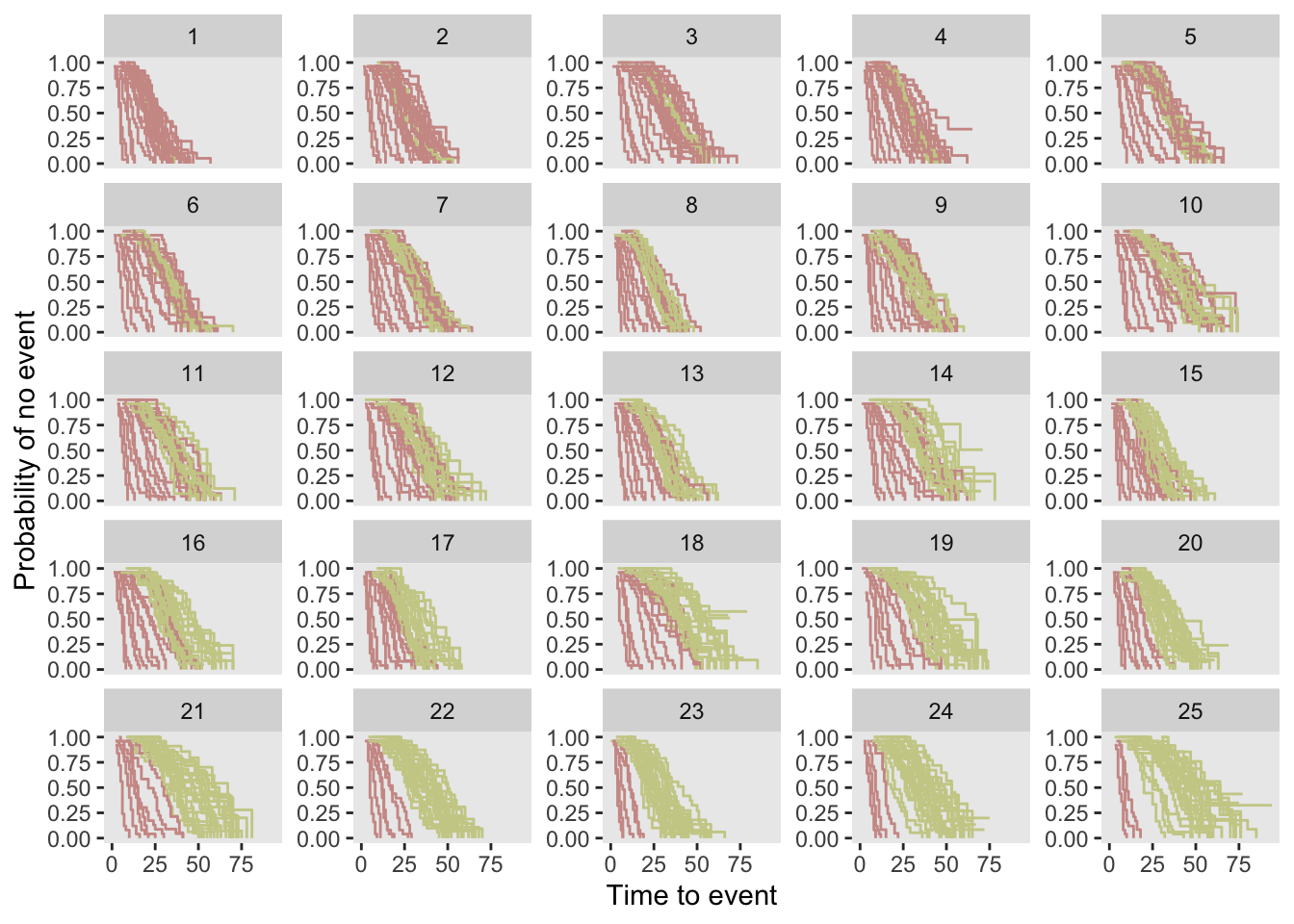
dd <- dd[, .(id, site, k, A, Y, event)]Here is a set of Kaplan-Meier plots for each site and enrollment period. When a site is in the intervention condition, the K-M curve is red. For simplicity, censoring is not shown, though about 20% of cases in this dataset are censored.
Model estimation
This model has quite a few components relative to the earlier models, but nothing is really new. There is a penalized spline for the effect of time and a random effect for each site. The primary parameter of interest is still .
For completeness, here is the model specification:
where
- : number of unique event times
- : number of spline basis functions
- is the set of individuals who experience an event at time .
- is the risk set at time , including all individuals who are still at risk at that time.
- is the number of events occurring at time .
- ranges from 0 to , iterating over the tied events.
- represents the average risk weight of individuals experiencing an event at :
- : binary indicator for treatment
- : value of the spline basis function for the observation
- : the second-difference matrix of the spline function
The parameters of the model are
- : treatment coefficient
- : spline coefficient for the spline basis function
- : cluster-specific random effect, where is the cluster of patient
- : the penalization term; this will not be estimated but provided by the user
The assumed prior distributions for and the random effects are:
And here is the implementation of the model in Stan:
stan_code <-
"
data {
int<lower=1> S; // Number of clusters
int<lower=1> K; // Number of covariates
int<lower=1> N_o; // Number of uncensored observations
array[N_o] int i_o; // Event times (sorted in decreasing order)
int<lower=1> N; // Number of total observations
matrix[N, K] x; // Covariates for all observations
array[N] int<lower=1,upper=S> s; // Cluster
// Spline-related data
int<lower=1> Q; // Number of basis functions
matrix[N, Q] B; // Spline basis matrix
matrix[N, Q] Q2_spline; // 2nd derivative for penalization
real<lower=0> lambda; // penalization term
array[N] int index;
int<lower=0> T; // Number of records as ties
int<lower=1> J; // Number of groups of ties
array[T] int t_grp; // Indicating tie group
array[T] int t_index; // Index in data set
vector[T] t_adj; // Adjustment for ties (efron)
}
parameters {
vector[K] beta; // Fixed effects for covariates
vector[S] b; // Random effects
real<lower=0> sigma_b; // SD of random effect
vector[Q] gamma; // Spline coefficients
}
model {
// Priors
beta ~ normal(0, 1);
// Random effects
b ~ normal(0, sigma_b);
sigma_b ~ normal(0, 0.5);
// Spline coefficients prior
gamma ~ normal(0, 2);
// Penalization term for spline second derivative
target += -lambda * sum(square(Q2_spline * gamma));
// Compute cumulative sum of exp(theta) in log space (more efficient)
vector[N] theta;
vector[N] log_sum_exp_theta;
vector[J] exp_theta_grp = rep_vector(0, J);
int first_in_grp;
// Calculate theta for each observation
for (i in 1:N) {
theta[i] = dot_product(x[i], beta) + dot_product(B[i], gamma) + b[s[i]];
}
// Compute cumulative sum of log(exp(theta)) from last to first observation
log_sum_exp_theta = rep_vector(0.0, N);
log_sum_exp_theta[N] = theta[N];
for (i in tail(sort_indices_desc(index), N-1)) {
log_sum_exp_theta[i] = log_sum_exp(theta[i], log_sum_exp_theta[i + 1]);
}
// Efron algorithm - adjusting cumulative sum for ties
for (i in 1:T) {
exp_theta_grp[t_grp[i]] += exp(theta[t_index[i]]);
}
for (i in 1:T) {
if (t_adj[i] == 0) {
first_in_grp = t_index[i];
}
log_sum_exp_theta[t_index[i]] =
log( exp(log_sum_exp_theta[first_in_grp]) - t_adj[i] * exp_theta_grp[t_grp[i]]);
}
// Likelihood for uncensored observations
for (n_o in 1:N_o) {
target += theta[i_o[n_o]] - log_sum_exp_theta[i_o[n_o]];
}
}
"Compiling the model:
stan_model <- cmdstan_model(write_stan_file(stan_code))Getting the data from R to Stan:
dx <- copy(dd)
setorder(dx, Y)
dx[, index := .I]
dx.obs <- dx[event == 1]
N_obs <- dx.obs[, .N]
i_obs <- dx.obs[, index]
N_all <- dx[, .N]
x_all <- data.frame(dx[, .(A)])
s_all <- dx[, site]
K <- ncol(x_all) # num covariates - in this case just A
S <- dx[, length(unique(site))]
# Spline-related info
n_knots <- 5
spline_degree <- 3
knot_dist <- 1/(n_knots + 1)
probs <- seq(knot_dist, 1 - knot_dist, by = knot_dist)
knots <- quantile(dx$k, probs = probs)
spline_basis <- bs(dx$k, knots = knots, degree = spline_degree, intercept = TRUE)
B <- as.matrix(spline_basis)
Q2 <- dbs(dx$k, knots = knots, degree = spline_degree, derivs = 2, intercept = TRUE)
Q2_spline <- as.matrix(Q2)
ties <- dx[, .N, keyby = Y][N>1, .(grp = .I, Y)]
ties <- merge(ties, dx, by = "Y")
ties <- ties[, order := 1:.N, keyby = grp][, .(grp, index)]
ties[, adj := 0:(.N-1)/.N, keyby = grp]
stan_data <- list(
S = S,
K = K,
N_o = N_obs,
i_o = i_obs,
N = N_all,
x = x_all,
s = s_all,
Q = ncol(B),
B = B,
Q2_spline = Q2_spline,
lambda = 0.15,
index = dx$index,
T = nrow(ties),
J = max(ties$grp),
t_grp = ties$grp,
t_index = ties$index,
t_adj = ties$adj
)Now we sample from the posterior - you can see that it takes quite a while to run, at least on my 2020 MacBook Pro M1 with 8GB RAM:
fit_mcmc <- stan_model$sample(
data = stan_data,
seed = 1234,
iter_warmup = 1000,
iter_sampling = 4000,
chains = 4,
parallel_chains = 4,
refresh = 0
)## Running MCMC with 4 parallel chains...## Chain 4 finished in 1847.8 seconds.
## Chain 1 finished in 2202.8 seconds.
## Chain 3 finished in 2311.8 seconds.
## Chain 2 finished in 2414.9 seconds.
##
## All 4 chains finished successfully.
## Mean chain execution time: 2194.3 seconds.
## Total execution time: 2415.3 seconds.fit_mcmc$summary(variables = c("beta", "sigma_b"))## # A tibble: 2 × 10
## variable mean median sd mad q5 q95 rhat ess_bulk ess_tail
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 beta[1] 0.815 0.815 0.0298 0.0298 0.767 0.865 1.00 3513. 5077.
## 2 sigma_b 0.543 0.535 0.0775 0.0739 0.432 0.683 1.00 3146. 5110.Estimating a “frequentist” random-effects model
After all that, it turns out you can just fit a frailty model with random effects for site and a spline for time period using the coxmme package. This is obviously much simpler then everything I have presented here.
frailty_model <- coxme(Surv(Y, event) ~ A + ns(k, df = 3) + (1 | site), data = dd)
summary(frailty_model)## Mixed effects coxme model
## Formula: Surv(Y, event) ~ A + ns(k, df = 3) + (1 | site)
## Data: dd
##
## events, n = 14989, 18750
##
## Random effects:
## group variable sd variance
## 1 site Intercept 0.5306841 0.2816256
## Chisq df p AIC BIC
## Integrated loglik 18038 5.00 0 18028 17990
## Penalized loglik 18185 27.85 0 18129 17917
##
## Fixed effects:
## coef exp(coef) se(coef) z p
## A 0.80966 2.24714 0.02959 27.36 <2e-16
## ns(k, df = 3)1 -2.71392 0.06628 0.04428 -61.29 <2e-16
## ns(k, df = 3)2 1.04004 2.82933 0.07851 13.25 <2e-16
## ns(k, df = 3)3 4.48430 88.61492 0.04729 94.83 <2e-16However, the advantage of the Bayesian model is its flexibility. For example, if you wanted to include site-specific spline curves—analogous to site-specific time effects—you could extend the Bayesian approach to do so. The current Bayesian model implements a study-wide time spline, but incorporating site-specific splines would be a natural extension. I initially hoped to implement site-specific splines using the mgcv package, but the models did not converge. I am quite confident that a Bayesian extension would, though it would likely require substantial computing resources. If someone wants me to try that, I certainly could, but for now, I’ll stop here.